A Quick Guide to Human Leukocyte Antigen (HLA) Typing Techniques
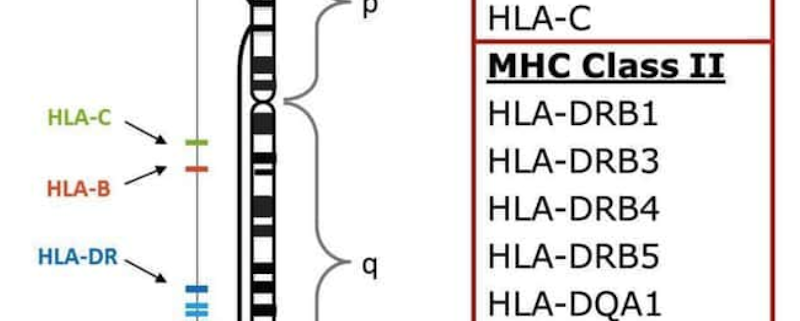
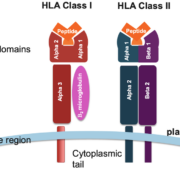
Human leukocyte antigens (HLAs) are proteins found on the surface of cells that play a vital role in the immune system. HLA typing is a critical process for determining compatibility between donors and recipients in organ transplantation, hematopoietic stem cell transplantation, and for understanding the genetic basis of many autoimmune diseases. This blog post delves into various HLA typing methods, highlighting their advantages, limitations, and applications in clinical and research settings.
Traditional Serological Techniques
Serological methods were the first techniques used for HLA typing. These methods rely on detecting HLA antigens using antibodies. The two primary serological techniques are complement-dependent cytotoxicity (CDC) and microlymphocytotoxicity (MLCT). While these methods are relatively simple and low-cost, they have several limitations, including low sensitivity and specificity, subjectivity in interpreting results, and inability to distinguish between closely related alleles.
Molecular Techniques
With advancements in molecular biology, HLA typing moved from serology to DNA-based techniques. Polymerase chain reaction (PCR) became the standard method for HLA typing, allowing for higher resolution and more accurate results. PCR-based methods include sequence-specific oligonucleotide (SSO) probes, sequence-specific primers (SSP), and sequence-based typing (SBT).
- SSO probes: This method uses fluorescent labeled oligonucleotide probes that hybridize to specific HLA sequences. The resulting fluorescence signals are detected and analyzed to determine the HLA type. SSO offers higher resolution than serology but may still be unable to distinguish between closely related alleles.
- SSP: This technique uses sequence-specific primers that amplify target HLA genes through PCR. The presence or absence of amplification products indicates the HLA type. While SSP provides high resolution, it can be labor-intensive and time-consuming, particularly for the highly polymorphic HLA system.
- SBT: This method involves PCR amplification of HLA genes followed by direct sequencing of the amplicons. SBT provides the highest resolution among molecular techniques, allowing for accurate identification of rare and novel alleles. However, it is more expensive and time-consuming than other molecular methods.
Next-Generation Sequencing (NGS)
The advent of next-generation sequencing (NGS) has revolutionized HLA typing, providing even higher resolution and accuracy. NGS enables the simultaneous sequencing of millions of DNA fragments, allowing for comprehensive and efficient HLA typing. NGS-based HLA typing can be performed using targeted gene panels or whole-genome sequencing. The major advantages of NGS include reduced labor and cost, improved accuracy, and the potential to identify novel alleles. However, challenges remain in data analysis and interpretation due to the complexity of the HLA system and the need for specialized bioinformatics tools.
Conclusion
HLA typing has evolved significantly over the years, from traditional serological methods to cutting-edge NGS technologies. Each technique has its own set of advantages and limitations, and the choice of method depends on the specific clinical or research application. As technology continues to advance, HLA typing is likely to become faster, more accurate, and more accessible, further improving our understanding of the complex relationship between HLA and the immune system.


 The Sequencing Center
The Sequencing Center
Leave a Reply
Want to join the discussion?Feel free to contribute!