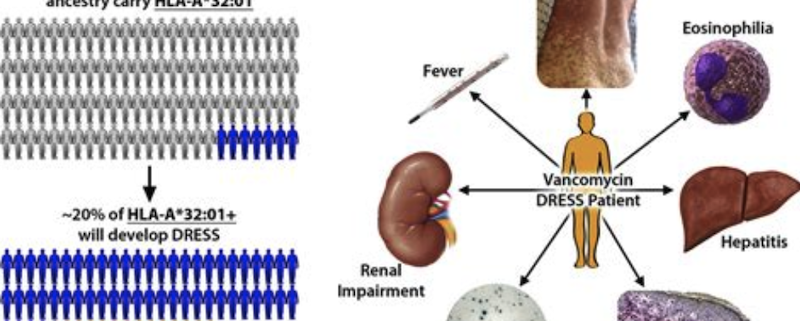
The HLA-A*32:01 Variant: A Focused Exploration for Immunology Experts
The Human Leukocyte Antigen (HLA) system is a cornerstone of the adaptive immune response, consisting of cell surface molecules encoded by the major histocompatibility complex (MHC) genes. The HLA-A gene, a member of the MHC class I family, is highly polymorphic, with thousands of allelic variants identified to date. The HLA-A32:01 variant has garnered particular attention due to its association with favorable outcomes in certain viral infections, including HIV. In this article, we will delve into the structural, functional, and clinical implications of the HLA-A32:01 variant, providing a comprehensive overview for fellow experts in the field.
Structural Characteristics of HLA-A*32:01
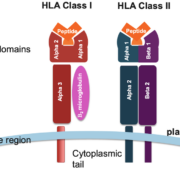
HLA-A32:01 encodes a class I MHC molecule that, like other HLA-A proteins, comprises an α heavy chain and a β2-microglobulin (β2m) light chain. The heavy chain consists of three extracellular domains (α1, α2, and α3), a transmembrane domain, and a cytoplasmic tail. The α1 and α2 domains fold together to form a peptide-binding groove that accommodates peptides derived from intracellular proteins. The α3 domain interacts with β2m, a non-polymorphic protein that contributes to the stability of the HLA-A32:01 molecule.
The peptide-binding groove of HLA-A32:01 possesses a unique set of amino acid residues that determine its peptide-binding specificity. These residues, found primarily in the α1 and α2 domains, govern the interactions between HLA-A32:01 and the peptides it presents, ultimately shaping the repertoire of antigens that can be recognized by CD8+ T cells.
Functional Implications of HLA-A*32:01 in Antigen Presentation
As an MHC class I molecule, HLA-A32:01 plays a critical role in presenting endogenous peptides to CD8+ T cells. The peptides, generated by proteasomal degradation of intracellular proteins, are transported into the endoplasmic reticulum (ER) by the transporter associated with antigen processing (TAP). Within the ER, peptides are loaded onto HLA-A32:01, forming a stable complex that is subsequently trafficked to the cell surface.
The HLA-A32:01/peptide complex can be recognized by the T cell receptor (TCR) on CD8+ T cells, initiating an immune response against cells harboring foreign antigens, such as viral peptides. The unique peptide-binding specificity of HLA-A32:01 influences the immune response by shaping the repertoire of antigens presented to CD8+ T cells, with potential implications for the host’s ability to combat specific pathogens.
HLA-A*32:01 and HIV Infection
HLA-A32:01 has been implicated in the immune control of HIV infection, with multiple studies demonstrating a correlation between HLA-A32:01 and slower disease progression. This association is thought to be mediated by the ability of HLA-A*32:01 to present conserved HIV epitopes that elicit a robust CD8+ T cell response, contributing to the control of viral replication.
One such HIV epitope is the Gag p24-derived peptide KIRLRPGGK (Gag 293-301), which is restricted by HLA-A32:01 and has been identified as a potential immune correlate of protection. CD8+ T cells targeting this epitope have been shown to exert strong antiviral activity, with HLA-A32:01-positive individuals displaying lower viral loads and higher CD4+ T cell counts compared to individuals lacking this allele.
The presentation of conserved HIV epitopes by HLA-A*32:01 may also contribute to a reduced likelihood of immune escape, as mutations within these epitopes could impair viral fitness. This phenomenon has been observed in the context of the Gag 293-301 epitope, where escape mutations have been associated with reduced viral replication capacity.
HLA-A*32:01 in Other Viral Infections
The role of HLA-A*32:01 in the immune response to other viral infections is an emerging area of research. Preliminary evidence suggests that HLA-A*32:01 may be associated with favorable outcomes in hepatitis C virus (HCV) infection, as it can present conserved HCV epitopes that elicit potent CD8+ T cell responses. Further studies are needed to elucidate the impact of HLA-A*32:01 on the immune control of HCV and other viral pathogens.
HLA-A*32:01 in Autoimmune Diseases
The potential role of HLA-A*32:01 in autoimmune diseases is less well-characterized compared to other HLA-A alleles. However, a few studies have reported associations between HLA-A*32:01 and certain autoimmune conditions. For instance, HLA-A*32:01 has been linked to an increased risk of idiopathic thrombocytopenic purpura (ITP), a rare autoimmune disorder characterized by the destruction of platelets. The exact mechanisms underlying this association remain to be elucidated.
HLA-A*32:01 Typing and Its Relevance in Transplantation
HLA-A*32:01 typing is an important aspect of donor-recipient matching in solid organ and hematopoietic stem cell transplantation. Given the role of HLA-A*32:01 in the immune response to viral infections, accurately identifying this allele may have implications for the management of post-transplant infections, particularly in patients with HIV or HCV.
Advanced HLA typing techniques, such as next-generation sequencing (NGS), enable high-resolution identification of HLA-A*32:01 and other alleles, facilitating optimal donor-recipient matching and potentially improving transplantation outcomes.
Conclusion
The HLA-A*32:01 variant is a fascinating subject in the field of immunology, with its unique peptide-binding specificity influencing the immune response to viral infections, particularly HIV. The association between HLA-A*32:01 and slower HIV disease progression highlights the importance of this allele in shaping the host’s ability to control viral replication. HLA typing can help elucidate the function of this allele.
Further research is needed to clarify the role of HLA-A*32:01 in other viral infections and autoimmune diseases, as well as its implications in transplantation. As our understanding of the HLA-A*32:01 variant deepens, it holds the potential to inform the development of novel therapeutic strategies aimed at enhancing the immune response to pathogens and improving transplantation outcomes.


 The Sequencing Center
The Sequencing Center
Leave a Reply
Want to join the discussion?Feel free to contribute!