HLA Typing and Gene Expression Profiling
Introduction
Human leukocyte antigen (HLA) typing has become an invaluable tool for understanding immune responses and investigating protein pathways associated with HLA genes. High-resolution HLA typing provides precise characterization of HLA alleles, enabling researchers to link specific HLA variants with disease susceptibility, vaccine efficacy, and transplantation outcomes[1][2]. Moreover, combining HLA typing data with other omics approaches like transcriptomics, proteomics, and immunopeptidomics further elucidates HLA-mediated biological processes[3]. This article reviews how integrating HLA typing with different omics datasets provides critical insights into HLA-related protein pathways and facilitates precision medicine applications.
HLA Typing Methodologies
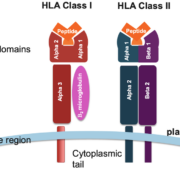
HLA typing methodologies have advanced considerably, transitioning from serological assays to next-generation sequencing (NGS) approaches that deliver nucleotide-level resolution. While serology-based methods only identify HLA antigen groups, sequence-specific oligonucleotide probes and sequencing determine precise HLA alleles defined by polymorphisms in antigen-binding grooves that alter peptide binding specificities[1][4]. High-throughput NGS platforms like Illumina and PacBio enable cost-effective and scalable HLA typing. Bioinformatics tools such as HLA-HD, HLAminer, and Omixon further facilitate analyzing NGS data, delivering accurate four-digit or six-digit resolution HLA typing[5].
Overall, progress in biochemistry, sequencing, and computing propels increasingly precise, rapid and affordable HLA typing with minimal ambiguities. State-of-the-art HLA typing provides the allele-level resolution necessary to investigate downstream impacts of HLA variants on disease and therapeutic outcomes.
Integrating HLA Typing With Other Omics
While HLA typing catalogs HLA alleles, integrating this genetic data with other omics approaches like transcriptomics and proteomics provides functional insights into HLA-mediated biology[3].
HLA Expression
Quantifying HLA RNA expression by RNA sequencing (RNA-seq) in tandem with HLA typing aids identification of expressed HLA alleles and enables digital HLA typing directly from transcriptomes. Comparing HLA expression across cell types, conditions, and diseases using public data from initiatives like the Cancer Cell Line Encyclopedia (CCLE) informs HLA functionality. Further, allele-specific HLA expression patterns can predict peptide binding capabilities more accurately than genetics alone.
Immunopeptidomics
Immunopeptidomics interrogates the repertoire of HLA-presented peptides (HLA ligandome) using mass spectrometry, providing insights into antigen processing and presentation pathways. Resources like IEDB catalog over 300,000 naturally presented HLA ligands across different alleles, cell types and diseases. Integrating large-scale HLA ligand datasets with HLA typing reveals allele-specific peptide binding motifs and pathways impacted by distinct HLA variants.
Multi-Omics
Combining HLA typing, transcriptomics, proteomics and metabolomics data using multi-omics techniques enables systems-level monitoring of HLA-mediated biology. Statistical and network modeling of multi-omics datasets elucidate HLA-regulated protein interactions and signaling cascades. Further, high-throughput multi-omics analysis facilitates identification of prognostic and diagnostic HLA biomarkers as well as therapeutic targets like neoantigens for immunotherapy.
Overall, complementing genetic HLA typing with functional omics data provides unprecedented insights into the downstream biological impacts of HLA alleles relevant to disease and medicine.
Wrapping Up
In summary, progress in high-resolution HLA typing combined with integrative omics approaches enables precise investigation of HLA-related protein pathways. HLA typing provides the cornerstone for elucidating HLA genetics while multi-omics analysis informs HLA-regulated protein interactions. Altogether, HLA typing and omics integration constitutes a powerful platform to decipher HLA-mediated immunity towards developing personalized diagnostics and therapeutics.
References[1] Shiina et al. J Immunol. 2009
[2] Gragert et al. HLA. 2013
[3] Boegel et al. HLA peptide affinity prediction. Springer, 2018[4] Eiz-Vesper et al. Front Immunol. 2012[5] Ozaki et al. HLA typing from RNA sequencing reads. Hum Immunol. 2018
Boegel et al. Genome Med. 2012 Bassani-Sternberg et al. Oncoimmunology. 2014
Abelin et al. Immunity. 2017 Caron et al. Mol Cell Proteomics. 2015 Vita et al. Nucleic Acids Res. 2019 Jurtz et al. Nat Biotech. 2017 Pearson et al. PLoS Comput Biol. 2016 Li et al. Brief Bioinform. 2013 Huang et al. Mol Cell Proteomics. 2018
Citations:
[1] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9162874
[2] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8195621
[3] https://pubs.acs.org/doi/10.1021/acs.jproteome.3c00491
[4] https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2023.1188381/full
[5] https://www.cell.com/cell-reports/pdf/S2211-1247(21)00681-1.pdf

 The Sequencing Center
The Sequencing Center
 The Sequencing Center
The Sequencing Center The Sequencing Center
The Sequencing Center The Sequencing Center
The Sequencing Center
Leave a Reply
Want to join the discussion?Feel free to contribute!