Using AlphaFold for Modeling HLA-Peptide Complexes in the Context of HLA Typing
The human leukocyte antigen (HLA) system plays a crucial role in the adaptive immune response, with HLA proteins presenting peptides derived from endogenous and exogenous proteins to T cells. Accurate modeling of the structural interactions between HLA proteins and their bound peptides is essential for understanding the molecular basis of immune recognition and has implications for vaccine design, transplantation, and immunotherapy.
Recent advances in protein structure prediction, particularly the development of AlphaFold by DeepMind, have opened up new possibilities for modeling HLA-peptide complexes. AlphaFold has demonstrated impressive performance in predicting the structures of individual proteins and protein-protein complexes, including HLA-peptide complexes.
One key advantage of AlphaFold is its ability to generate accurate models even in the absence of close structural templates. This is particularly relevant for HLA proteins, which exhibit high sequence diversity across different alleles. Traditional structure prediction methods that rely heavily on template-based modeling may struggle to capture the structural variations among different HLA alleles.
To leverage AlphaFold for modeling HLA-peptide complexes, researchers have developed specialized pipelines that incorporate HLA typing information. These pipelines typically involve the following steps:
- HLA typing: High-resolution HLA typing is performed using methods such as PCR sequence-specific primers (SSPs) or next-generation sequencing.
- Peptide selection: Peptides of interest, such as those derived from viral antigens or tumor neoantigens, are identified based on their potential to bind specific HLA alleles.
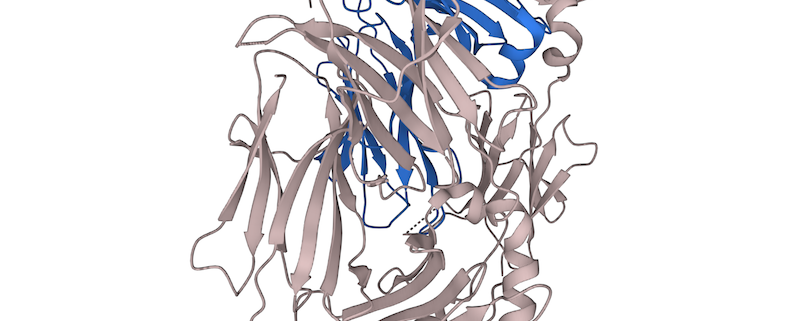
- AlphaFold modeling: The sequences of the HLA protein and the selected peptide are provided as input to AlphaFold, along with any available structural templates. AlphaFold generates a set of predicted models for the HLA-peptide complex.
- Model evaluation: The predicted models are assessed based on various metrics, such as the predicted local distance difference test (pLDDT) score and the root-mean-square deviation (RMSD) from known structures. Models with high confidence scores are selected for further analysis.
The resulting AlphaFold-generated models can provide valuable insights into the structural basis of HLA-peptide interactions. For example, they can reveal key residues involved in peptide binding, help identify potential T cell epitopes, and guide the design of peptide-based vaccines or immunotherapies.
However, it is important to note that AlphaFold-based modeling of HLA-peptide complexes is not without limitations. The accuracy of the predicted models may vary depending on the specific HLA allele and peptide sequence, and experimental validation is still necessary to confirm the predicted interactions. Additionally, AlphaFold does not explicitly model the dynamics and flexibility of HLA-peptide complexes, which may be important for understanding their biological function.
Despite these limitations, the integration of AlphaFold with HLA typing represents a promising approach for advancing our understanding of HLA-peptide interactions and their role in the immune response. As the field continues to evolve, we can expect further refinements in the modeling pipelines and increased synergy between computational predictions and experimental validation.

 The Sequencing Center
The Sequencing Center
Leave a Reply
Want to join the discussion?Feel free to contribute!