What is index dropout?
In Illumina sequencing, indexing (barcoding) is the process of adding unique tags to each read in a specific sample. The indexing step is performed during library preparation after the genomic fragmentation step. After fragmentation, each read in a sample can be tagged to uniquely associate that read with a specific sample. Once all reads in a sample have been indexed they can be pooled (multiplexed) with other reads from other samples and sequenced together in a single run. When sequencing is complete a binning algorithm separates all the pooled reads into unique bins according to their unique indexes. Thus, each bin contains only the reads for a single specific sample.
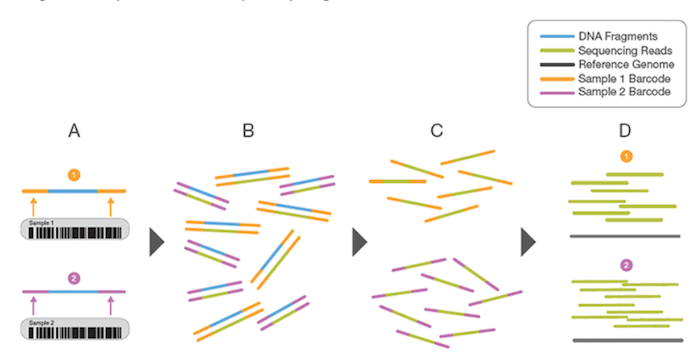
Figure 1.
Fig. 1 shows a high-level conceptual view of sample indexing (barcoding).
A) Two representative DNA fragments from two unique samples, each attached to a specific index (barcode) sequence, that identifies the sample from which it originated.
B) Libraries for each sample are pooled and sequenced in parallel. Each new read contains both the fragment sequence and its sample-identifying index (barcode).
C) Index (barcode) sequences are used to de-multiplex or differentiate reads from each sample.
D) Each set of reads is aligned to a reference sequence.
Indexing (barcoding) sample reads is an effective method for differentiating reads from two or more pooled samples. Indexing allows us to pool multiple samples in a single sequencing run, often up to 48 samples in a typical HLA run, resulting in very significant cost savings. Pooling indexed samples also improves turnaround time, results in more efficient use of library prep reagents and improves the scheduling of laboratory staff activities.
In some fairly rare cases we may see index dropout for a specific sample in a sequencing run. Index dropout manifests itself when the binning algorithm assigns a very small number of reads to a sample at the conclusion of a run. For example, if a sample has, say, 1M reads assigned to it (based on the library prep procedures and the total number of pooled samples), but the binning algorithm only assigns, say, 10K reads to the sample, then we call this issue index dropout. Because there is such a small number of reads assigned to the final sample, it cannot be used for any meaningful HLA typing analysis. Typically, in this case, we would attempt a second sequencing run with the sample, perhaps after performing DNA cleanup and/or DNA concentration protocols on the sample and possibly choosing a different index sequence from the first run.
Note that the root cause for index dropout is not always clear. There may be several factors that contribute to it including possible contaminants in the original sample that inhibit PCR amplification, insufficient DNA concentration before starting library prep, some kind of interference between the index oligonucleotide sequence and adapter sequences or the read sequences themselves, insufficient nucleotide diversity that inhibits downstream basecalling, or other unknown factors. In some cases we simply don’t know what causes index dropout and have to run the sample again.