What is index hopping?
Index hopping occurs when index (barcode) sequences initially assigned to a specific sample are incorrectly assigned to other sample(s) in a pool of samples. The result of index hopping is the incorrect assignment of sample reads (DNA fragments) in pooled samples. Index hopping only applies to pooled or multiplexed sequencing runs, i.e. when two or more samples are combined in a single run. This note only refers to index hopping in Illumina sequencing instruments and the GenDx HLA library preparation protocol. Note that index hopping is not unique to the Illumina platform or GenDx protocols and is seen in other sequencing platforms and protocols.
Index Hopping
Fig. 1
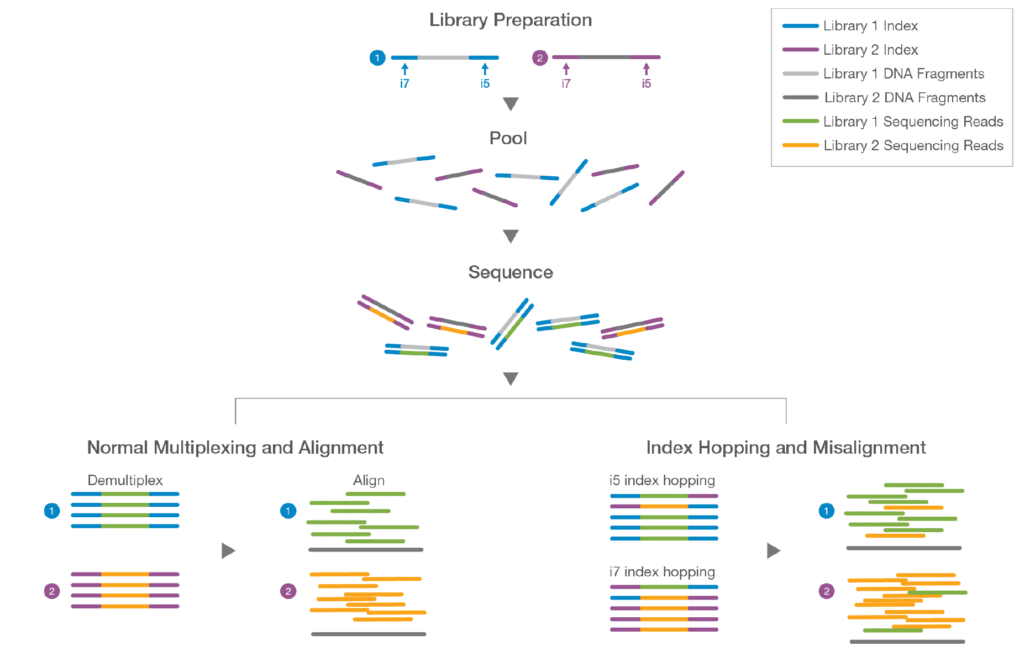
Fig. 1 shows schematically the library preparation process, sample pooling, sequencing and indexing for two individual samples (libraries). Sample 1 DNA fragments and sample 2 DNA fragments are shown in light grey and dark grey, respectively. Sample 1 DNA fragments are flanked by unique Illumina i5 and i7 indexes (blue); Sample 2 DNA fragments are also flanked by unique Illumina i5 and i7 indexes (purple). The blue i5/i7 index sequences are different from the purple i5/i7 index sequences, thus, the sample 1 and sample 2 DNA fragments can be uniquely identified by their respective unique index sequences.
During the library prep process, once each sample has been uniquely indexed, the two samples can be safely pooled together in a single combined (multiplexed) sample. The pooled samples are sequenced on an Illumina sequencer (i.e. MiniSeq).
When sequencing is complete a demultiplexing algorithm separates sample reads into unique bins according to their library indexing strategy – sample 1 reads (green) and their respective i5/i7 indexes go into one bin and sample 2 reads (orange) and their respective i5/i7 indexes go into another bin. Once the samples are safely binned, the demultiplexing algorithm then also strips out index sequences from the sample reads. The sample reads are then available for downstream data analysis, such as alignment to a reference genome.
However, index hopping can introduce noise in the signal. There is a low probability that some i5 indexes from sample 1 (blue) can wind up on sample 2 reads or i5 indexes from sample 2 (purple) can wind up on sample 1 reads. Likewise, some i7 indexes from sample 1 (blue) can wind up on sample 2 reads or i7 indexes from sample 2 (purple) can wind up on sample 1 reads. In this manner the demultiplexer could assign sample 1 reads (green) to the sample 2 bin or assign sample 2 reads (orange) to the sample 1 bin. This erroneous assignment of reads would then interfere with the correct interpretation of results in downstream bioinformatic data analysis.
Mitigation Strategies for Index Hopping
The primary culprit that causes index hopping seems to be the presence of free adapters in samples during the library prep process, especially during the pooling or multiplexing steps. Apparently, free floating, unligated adapter sequences can hybridize with the wrong i5 or i7 index sequence, thus causing the wrong index to “hop” over to a different sample read. The elimination of free adapters is of paramount importance during library prep.
Here are some mitigation strategies and best practices from Illumina and GenDx to minimize or eliminate index hopping:
- Library purity. There are several quality control cleanup steps in our HLA library prep protocols. Each cleanup step removes free adapters from samples and the combined cleanup steps should remove essentially all adapters.
- Library storage. Sample libraries should be stored at -20o C. Libraries should not be stored at 4o C or room temperature. We store all libraries at -20o C.
- Library pooling. Libraries should be pooled just prior to sequencing. We pool sample libraries shortly before sequencing, as specified in our standard operating procedures.
- Library prep methods. Illumina research has shown that PCR-free library prep protocols significantly increase the likelihood of index hopping. Our GenDx HLA library prep procedures include at least three PCR steps, which should minimize index hopping.
- Patterned flow cells. Some Illumina instruments use patterned flow cells. Index hopping occurs at a somewhat higher rate in these types of flow cells. We use Illumina instruments (i.e. MiniSeq) that use non-patterned flow cells, hence, index hopping should be reduced on our instruments.
- Use Unique Dual Indexing (UDI). UDI is a combinatorial method of generating unique Illumina i5 and i7 index sequences. For practical purposes, UDI entirely eliminates index hopping. GenDx uses index adapter plates with UDI combinations of i5/i7 indexes. This method of pooling and indexing should eliminate index hopping.
Illumina research has shown that if index hopping does occur, it generally affects about 0.1 – 0.3% of reads in a pooled multiplexed sequencing run. With no mitigation strategies index hopping should be a fairly minor issue. Nevertheless, by using some of the mitigation strategies mentioned here, we believe index hopping is likely eliminated in our HLA protocols.