How do we quantitate DNA?
DNA concentration is key. In our HLA sequencing protocols, one of the first, and most important, steps is to accurately determine the concentration of dsDNA (double-stranded DNA). This is a critical QC (quality control) check that affects all downstream steps in our HLA protocols. Considering that there are more than 100+ steps in our HLA protocols, we must ensure that we start with sufficient DNA concentration. To do this we use Qubit fluorometric methods for DNA quantitation.
Fig. 1
As shown in Fig. 1, the Qubit fluorometer is a powerful instrument used for quantitating various molecular species including dsDNA, RNA, oligonucleotides (i.e. ssDNA) and proteins. This particular model (Qubit 3) includes an Ion Sphere assay for use with the Ion Personal Genome Machine Sequencer and a Fluorometer mode that allows the instrument to be used as a mini-fluorometer. In this note we’re only interested in the dsDNA quantitation mode.
This Qubit model supports several types of quantitation assays, including the dsDNA BR (broad-range) Assay, the dsDNA HS (high-sensitivity) Assay, the ssDNA (single-strand) Assay, the RNA BR (broad-range) Assay, the RNA HS (high-sensitivity) Assay, the RNA XR (extended range) Assay, the RNA IQ Assay (for degraded RNA) and the Protein BR (broad-range) Assay.
We’ll only consider the dsDNA BR Assay, which is used to quantitate double-stranded DNA for our HLA protocols. This assay accuratly quantifies DNA concentrations in the range 100 pg/uL – 1,000 ng/uL. The assay is performed at room temperature and is highly tolerant of common sample contaminants such as RNA, free nucleotides, salts, solvents, detergents and proteins.
The Qubit DNA quantitation protocol consists of two main steps: 1) running an instrument and assay calibration check and 2) calculating the sample DNA concentration. The calibration reagents and buffers are temperature sensitive so it’s important to run a new calibration check at the beginning of each BR assay.
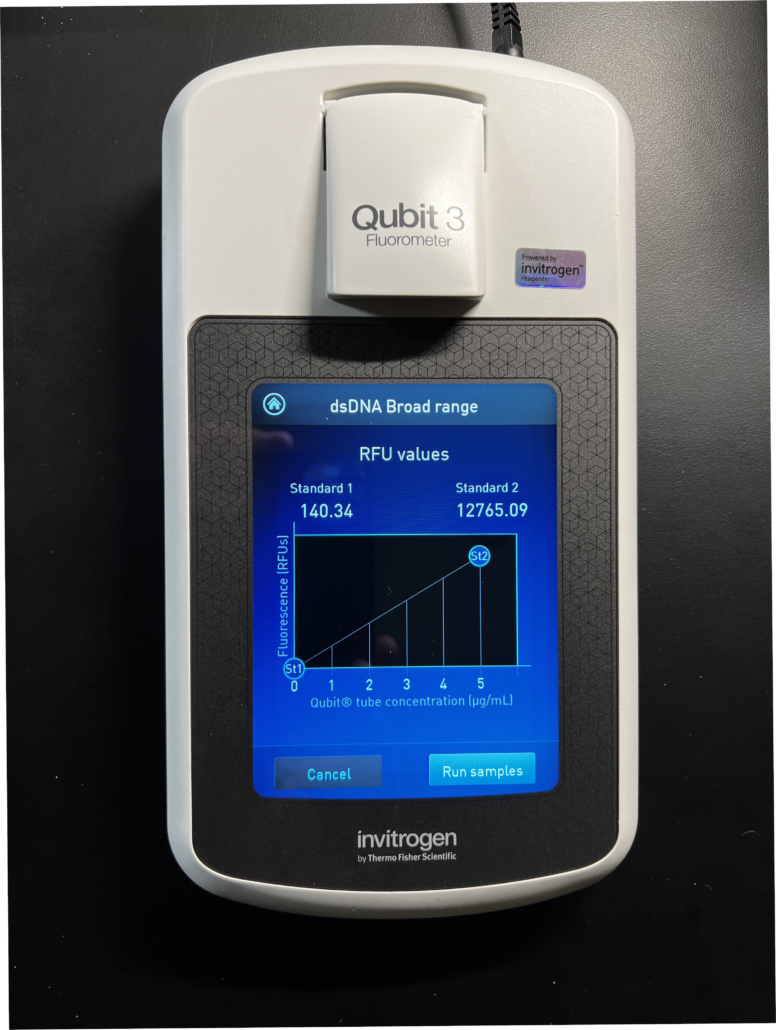
Fig. 2
To calibrate the instrument we prepare two standard reagents, aptly named “Standard 1” and “Standard 2”. As shown in Fig. 2, the fluorescence signals from these two standards generate a linear calibration curve measured in RFU’s (relative fluorescence units). Typically, Standard 1 displays around 100 RFU’s and Standard 2 displays around 10,000 RFU’s, however, there is significant and acceptable variability around these values. The calibration curve shown in Fig. 2 is very typical for a DNA quantitation run.
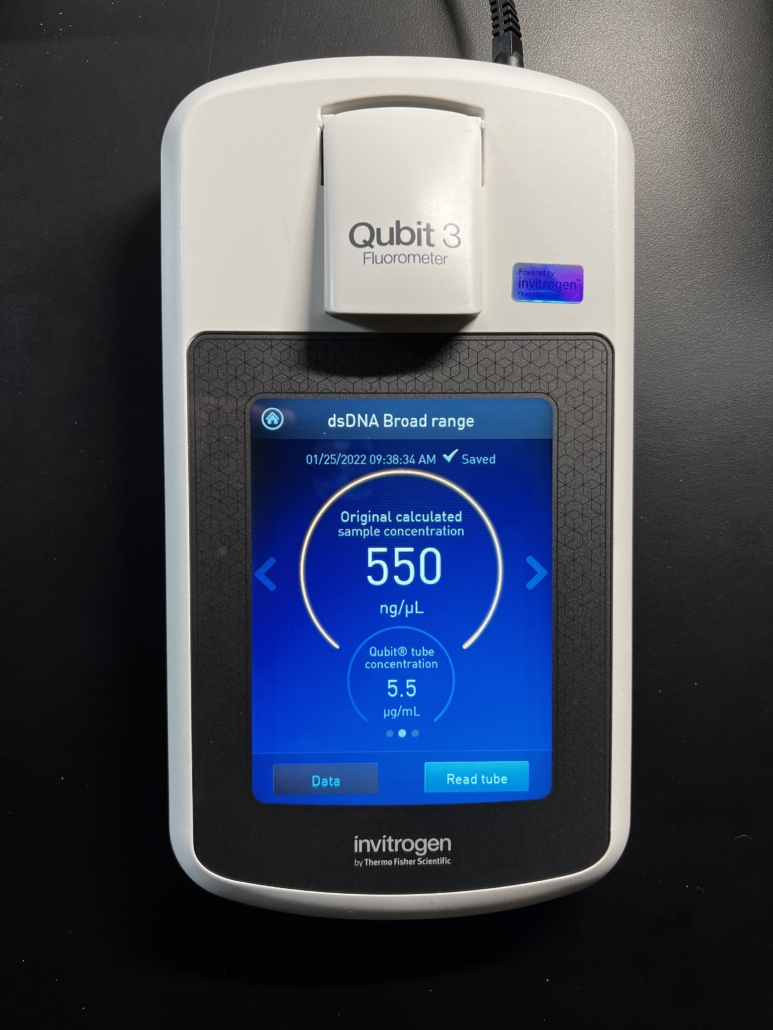
Fig. 3
After calibration we test one or more samples to determine their DNA concentration. As shown in Fig. 3, this particular sample registered a DNA concentration of 550 ng/uL. This is excellent concentration for our HLA protocols and in fact will have to be diluted down to about 30 ng/uL – 100 ng/uL, the optimal range, before starting the HLA procedures. Note that this metric of 550 ng/uL pertains only to dsDNA – it does not represent the concentration of any other molecular species that may be present in the sample. Thus, it is an extremely accurate representation of the amount of dsDNA available for sequencing. This is in contrast to spectrophotometric methods (i.e. Nanodrop) that are commonly used to determine DNA concentration. See our Knowledge Base article that compares and contrasts these two methods.
Accurately measuring dsDNA concentration is one of the most important QC checks that we perform for HLA sequencing. And Qubit fluorometric methods are indispensible for this task.