Leveraging AI and ML in HLA Typing: A New Frontier in Health Science
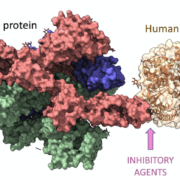
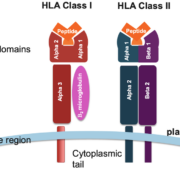
Our understanding of human health is constantly evolving, and at the cutting edge of this expansion is the increasingly critical role of Human Leukocyte Antigen (HLA) typing. This method, key in immunogenetic research, involves identifying specific variations in the human genome that can influence immune response. A dauntingly complex task, HLA typing is being streamlined by advancements in Artificial Intelligence (AI) and Machine Learning (ML). These technologies are transforming the way we interpret HLA sequencing results, revolutionizing diverse sectors from organ transplantation to disease susceptibility and pharmacogenomics.
Organ transplantation stands as one of the most critical areas in which AI and ML have proven transformative. The successful pairing of donor and recipient relies heavily on HLA typing, as matching HLA alleles can significantly reduce the risk of organ rejection. However, the diversity and complexity of HLA alleles present a challenge. To address this, AI-based algorithms have been developed to efficiently analyze HLA sequencing data. These tools predict potential matches, thereby optimizing the selection process and increasing the chances of successful transplantation. With such technology, we are moving towards a future where the trial and error in organ transplantation are substantially reduced.
In the realm of disease susceptibility, AI and ML have further enhanced the role of HLA typing. Complex diseases such as diabetes, rheumatoid arthritis, and certain types of cancer have been linked to specific HLA alleles. To navigate this complexity, researchers are utilizing machine learning models to identify patterns in the massive volumes of HLA data. These patterns help pinpoint specific HLA alleles linked to particular diseases, thus enabling early detection and personalized treatment plans. As such, AI and ML offer invaluable tools in preempting and managing a variety of health conditions.
For example the study titled “A deep learning method for HLA imputation and trans-ethnic MHC fine-mapping of type 1 diabetes”1 used artificial intelligence in HLA typing. The study developed a deep learning method called DEEP*HLA for imputing HLA genotypes. This method was found to be significantly superior for low-frequency and rare alleles. DEEP*HLA was less dependent on distance-dependent linkage disequilibrium decay of the target alleles. The researchers applied DEEP*HLA to Type 1 diabetes GWAS data from BioBank Japan (n = 62,387) and UK Biobank (n = 354,459), and successfully disentangled independently associated Class I and II HLA variants with shared risk among diverse populations.
Shifting focus to pharmacogenomics, the integration of AI and ML in HLA typing presents compelling possibilities. Pharmacogenomics studies how genes influence a person’s response to drugs. Given the role of HLA in the immune response, it has been identified as a significant factor in drug reactions. AI and ML are being used to parse through copious amounts of HLA typing data to predict an individual’s potential reaction to a specific drug, thereby avoiding adverse reactions and enhancing therapeutic efficiency.
In summary, the incorporation of AI and ML in HLA typing heralds a paradigm shift in medical science. The application of these technologies not only expedites HLA sequencing but also enhances its accuracy, thus facilitating greater success in organ transplantation, improved understanding of disease susceptibility, and a more personalized approach in pharmacogenomics. While challenges remain, the potential of AI and ML to revolutionize these areas is evident, with far-reaching implications for human health. As we continue to refine these technologies, the future of HLA typing and its associated fields looks increasingly promising.
1Naito, T., Suzuki, K., Hirata, J. et al. A deep learning method for HLA imputation and trans-ethnic MHC fine-mapping of type 1 diabetes. Nat Commun 12, 1639 (2021).



 The Sequencing Center
The Sequencing Center
 The Sequencing Center
The Sequencing Center
Leave a Reply
Want to join the discussion?Feel free to contribute!