How to perform an NCBI nucleotide BLAST search
We often use NCBI nucleotide Blast queries to identify DNA sequences derived from Next Generation Sequencing (NGS) runs. This brief note describes how to use the NCBI Blast algorithm and website with NGS DNA datasets.
Create an NCBI Account
Before starting an NCBI Blast search, you may want to create an NCBI account so that you can save and retrieve your Blast queries at a later date. To create an account, go to the NCBI Blast home page, click “Sign in to NCBI” in the upper right-hand corner, click “Register for an NCBI account” in the lower left-hand corner and follow the instructions therein. After creating an account, you can log in to the account on the NCBI Blast home page.
Required Blast Query Files
We can supply the appropriate files for Blast queries from your sequencing runs. Typically these will be in one of the following file formats:
- Short-reads, i.e. DNA fragments
- Relatively “small” genomes, i.e. bacteriophage, plasmids, expression vectors, etc.
- De novo assembled contigs or scaffolds
- FASTA file format
How to Perform a Nucleotide Blast Search
The general procedure for a nucleotide Blast search is as follows:
1. Navigate to the NCBI Blast home page.
2. Click on the “Nucleotide BLAST” box.
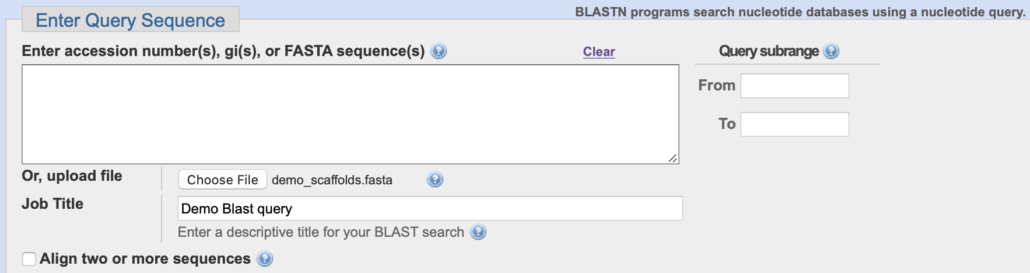
3. In the “Enter Query Sequence” selection box, click “Choose File”.
Navigate to a query file on your desktop and click “Choose”.
The query file name should appear next to “Choose File” (i.e. “demo_scaffolds.fasta” in the example below).
In the “Job Title” field, enter a unique name for the Blast query (i.e. “Demo Blast query” in the example below).
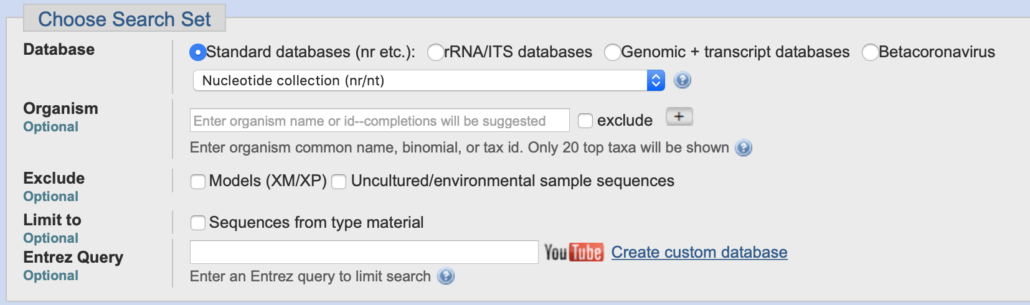
4. In the “Choose Search Set” selection box, click “Standard databases”.
In the drop-down list choose “Nucleotide collection (nr/nt)”.
Leave all other fields in their default state.
5. In the “Program Selection” selection box, choose “Highly similar sequences (megablast)”.
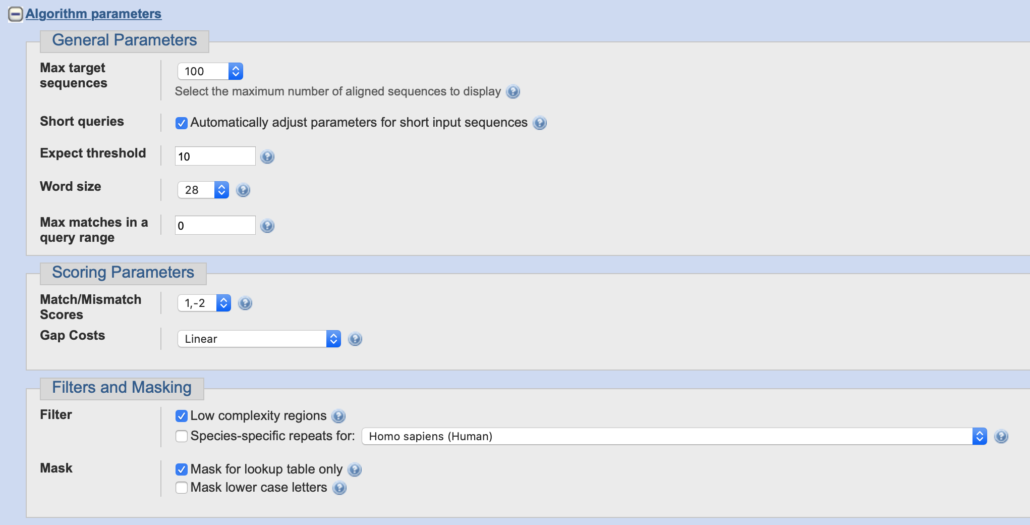
6. Click the plus (“+”) sign by “Algorithm parameters” to expand the parameters list. Ensure that all of the default settings are configured correctly. (You can click “Restore default search parameters” in the upper right-hand corner to reset the default settings). Click the minus (“-“) sign by “Algorithm parameters” to close the parameters list.
7. Click the “BLAST” button to initiate a Blast search.
How to Read Blast Query Results
After some period of time, the Blast algorithm will display a results page.
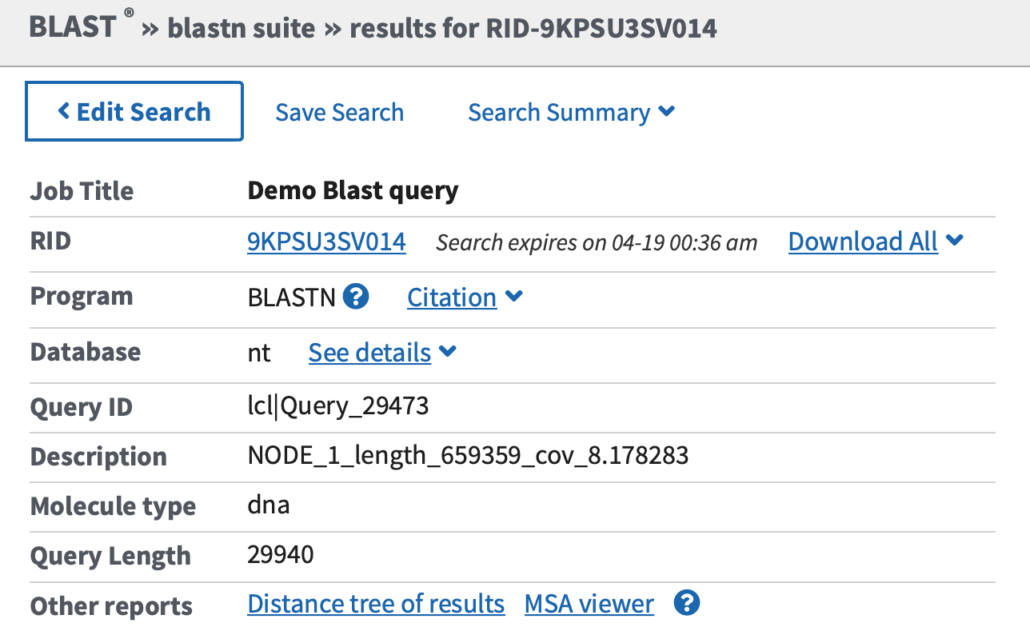
Fig. 1
As shown in Fig. 1, the top of the search results page displays metadata about the query. “RID” is the Blast search Request ID number. You can use RID to retrieve this Blast search at a later date. Note that the search expires after 48 hours so you may want to use “Save Search” if you intend to keep the results for future reference.
Fig. 2
Fig. 2 shows the results of a Saved Blast query. You can view or download the query from this page.
Fig. 3
As shown in Fig. 3, the Blast query alignments are displayed in rank order according to their “Max Score”. For this example, the top hit from the Demo scaffold sequence was Escherichia coli strain 2011C-3911 with Accession #CP015240.1.
Fig. 4
Clicking the Description name for one of the Blast hits will display a sequence alignment of the query sequence vs. the NCBI nucleotide database sequences, as shown in Fig. 4. This result includes the Karlin-Altschul Expect value, the percent identity of the alignment, the number of gaps in the alignment, and other pertinent information.
Additional Resources
There is a wealth of information generated from NCBI Blast queries, more than we can cover in this brief note. Here are some resources that will help navigate the NCBI Blast nucleotide site: