How do you read HLA reports?
We deliver detailed HLA reports to clients immediately after their HLA sequencing runs are complete. The reports include information about HLA allele typing calls, allele ambiguities, and relevant metadata.
HLA Loci
The full set of HLA loci available for sequencing and typing is:
- HLA-A
- HLA-B
- HLA-C
- DPA1
- DPB1
- DQA1
- DQB1
- DRB1
- DRB3
- DRB4
- DRB5
- HLA-G (optional)
The exact set of loci displayed in an HLA report will depend on which loci were chosen for sequencing. We can sequence individual loci, a subset of the 11-loci, or the full 11-loci. We can also sequence the HLA-G allele upon request.
Example HLA Report
We’ll review here the major elements in a sample HLA report.
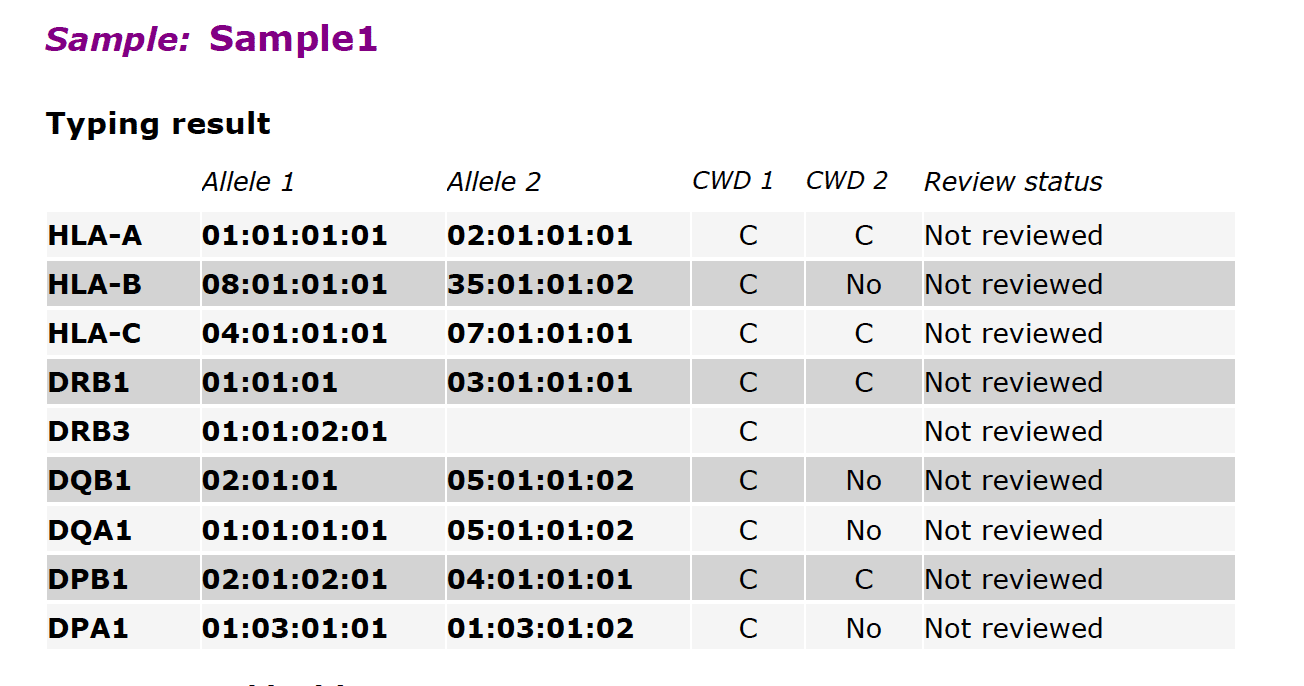
Fig. 1
Fig. 1 shows an overview of typing results for all loci in a single sample. For each locus, the typing call for “Allele 1” and “Allele 2” is shown. By default, these are displayed to 4-digit accuracy. If desired, we can display 1-digit, 2-digit, or 3-digit accuracy instead of 4-digit accuracy.
The CWD (Common and Well-Documented) status of each allele is shown next, as follows:
- C = Common
- WD = Well Documented
- No = Not available in the IMGT/HLA CWD list
CWD alleles are incorporated in the IMGT/HLA database and are updated periodically as immunogenetics researchers discover additional alleles. If an allele, and therefore its corresponding CWD status, is missing in the report (blank field), or the field contains “No typing result available”, this indicates that the allele itself is missing in the sample or in the subject’s genome. This most often occurs in HLA genes DRB3, DRB4, and DRB5. These genes are frequently absent in familial or ethnic cohorts.
The Review status can be one of four states:
- Not reviewed: shows typing results of loci that have not been reviewed
- Rejected: shows typing results of loci that have been rejected
- First reviewer approved: shows typing results of loci that have been approved by the first reviewer
- Second reviewer approved: shows typing results of loci that have been approved by the second reviewer
For most HLA runs, the Sequencing Center does not review and approve/reject the HLA loci typing results. This function is typically assigned to designated reviewers in the client organization. So in most reports delivered by The Center, the Review status will be “Not reviewed”. If the Review status appears as “Accepted (1st rev.)”, this indicates that The Sequencing Center staff checked the sequencing quality control metrics and considered them acceptable.
Just below the Typing Result table, there may be a footnote with the following format:
Exon+ mismatches: xx
where “xx” is some integer value. For example, it may read “Exon+ mismatches: 1”. The HLA allele typing algorithm performs sequence alignments and variant calls of sample data against the IMGT/HLA database and attempts to make a statistical estimate of the best matching genotype between the sample data and IMGT database. It may flag some exon base call mismatches between the sample and database as “Exon+ mismatches”. In the example above the algorithm flagged a single exon base call as a mismatch. The HLA typing algorithm uses Exon mismatches, along with many other parameters, to determine the best matching genotype for each HLA gene.
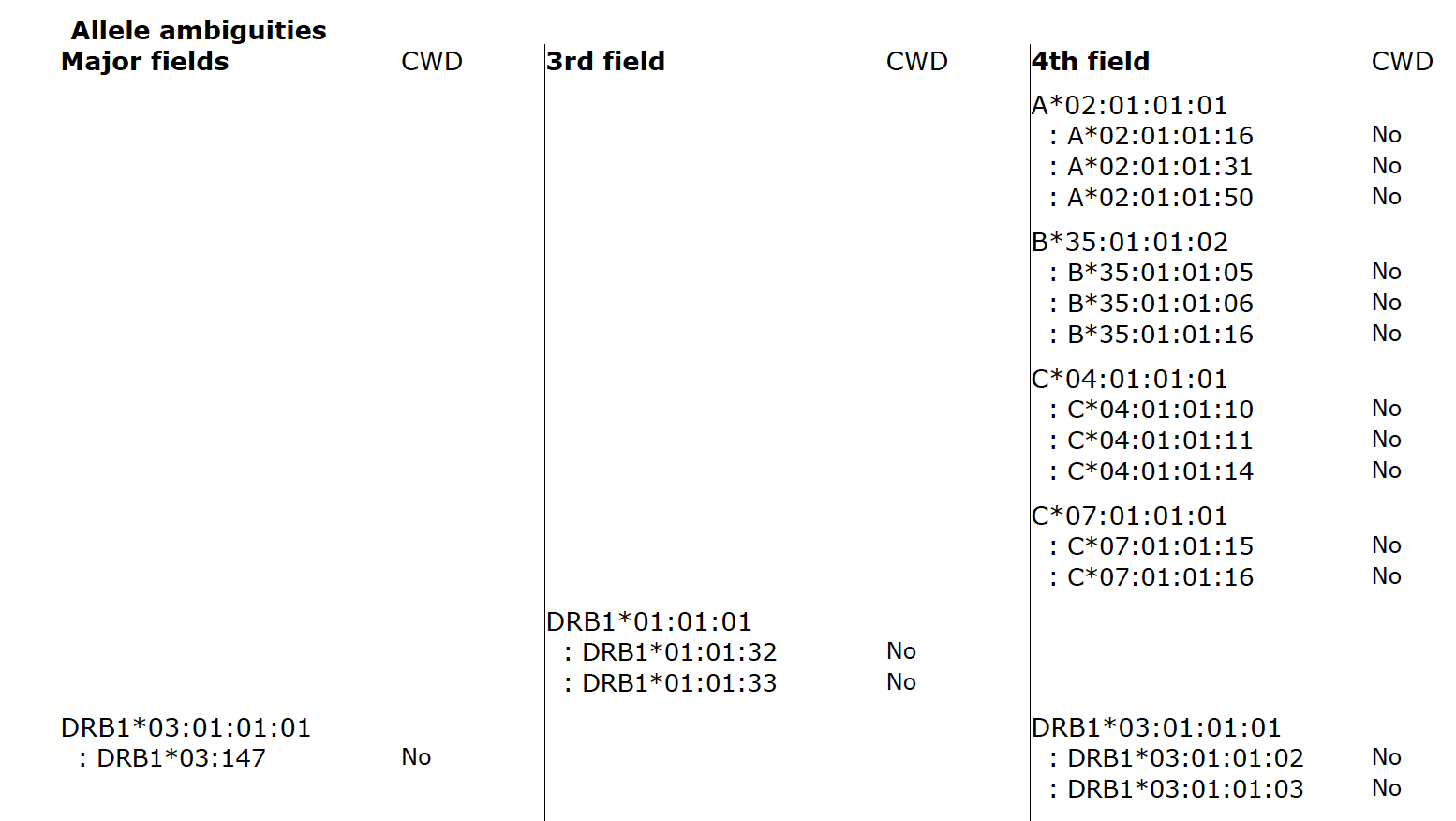
Fig. 2
Fig. 2 shows allele ambiguities in the major fields (first field, second field), third field, and fourth field. If typing ambiguities are present the report will list each of them, including their CWD status.
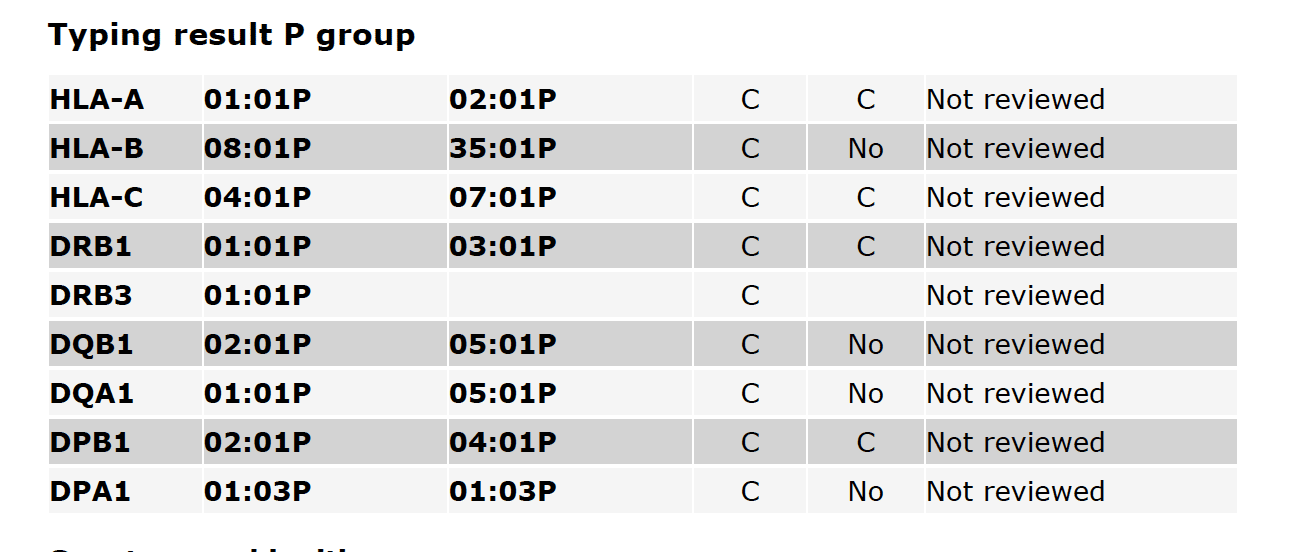
Fig. 3
Fig. 3 shows P-group typing results, including the CWD status of alleles and the Reviewer state. A more detailed description of P-group typing is available on the HLA Nomenclature site.
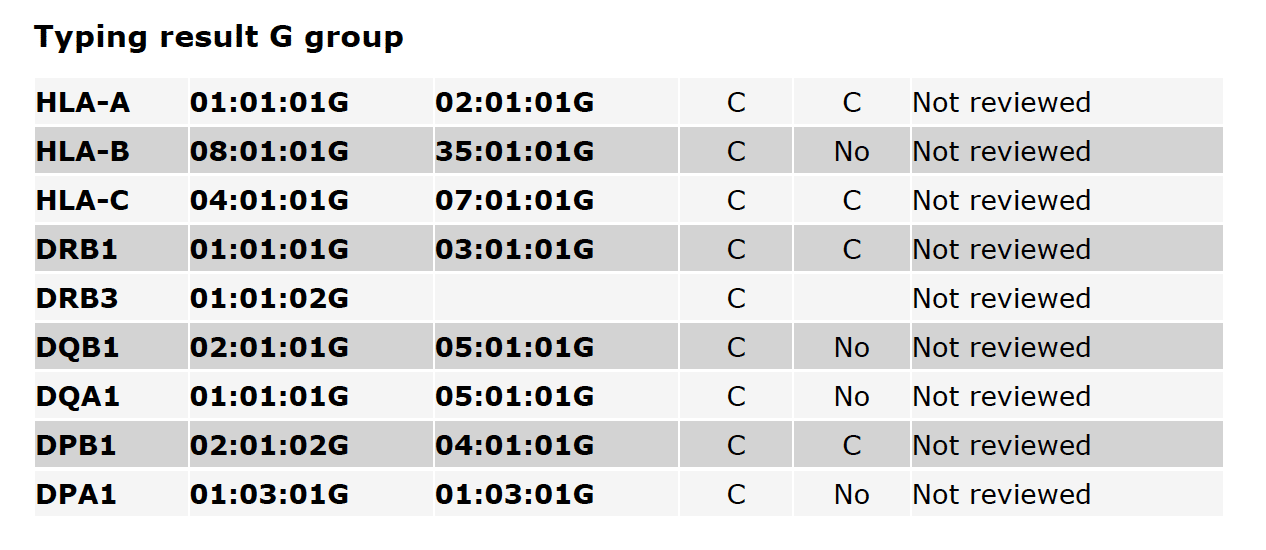
Fig. 4
Fig. 4 shows G-group typing results, including the CWD status of alleles and the Reviewer state. A more detailed description of G-group typing is available on the HLA Nomenclature site.
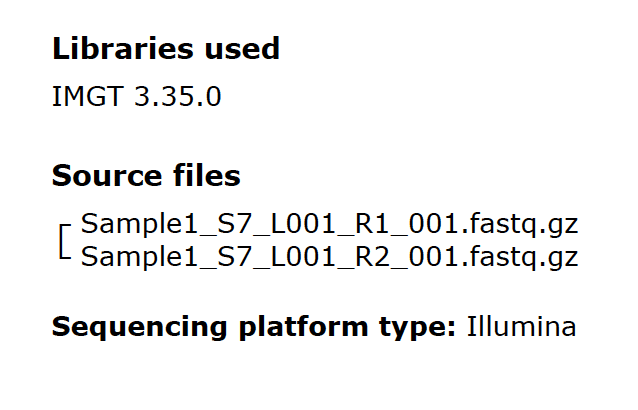
Fig. 5
Fig. 5 shows some metadata for each sample in a report. The IMGT database library is the most current version available at the time of the sequencing run. The source files for HLA typing are gzip compressed FastQ files. The file naming convention includes the sample name plus some information appended by the Illumina sequencers. The sequencing platform is also identified in the report.