What is the IMGT/HLA database?
IMGT Database
The IPD-IMGT/HLA database contains genetic sequence and allele data for the human major histocompatibility complex (MHC). This database is foundational for many HLA Typing algorithms and Next-Generation sequencing projects. It includes the most commonly investigated MHC Class I and Class II genes and alleles, such as:
Class I
Class II
Database Tools
The database includes a variety of query and analysis tools for interrogating HLA Typing calls. These include:
- Protein similarity search
- Nucleotide similarity search
- Various query tools
- Pairwise sequence alignment tools
- Multiple sequence alignment tools
- Allele query tool
- Ambiguous allele search tool
Database Growth
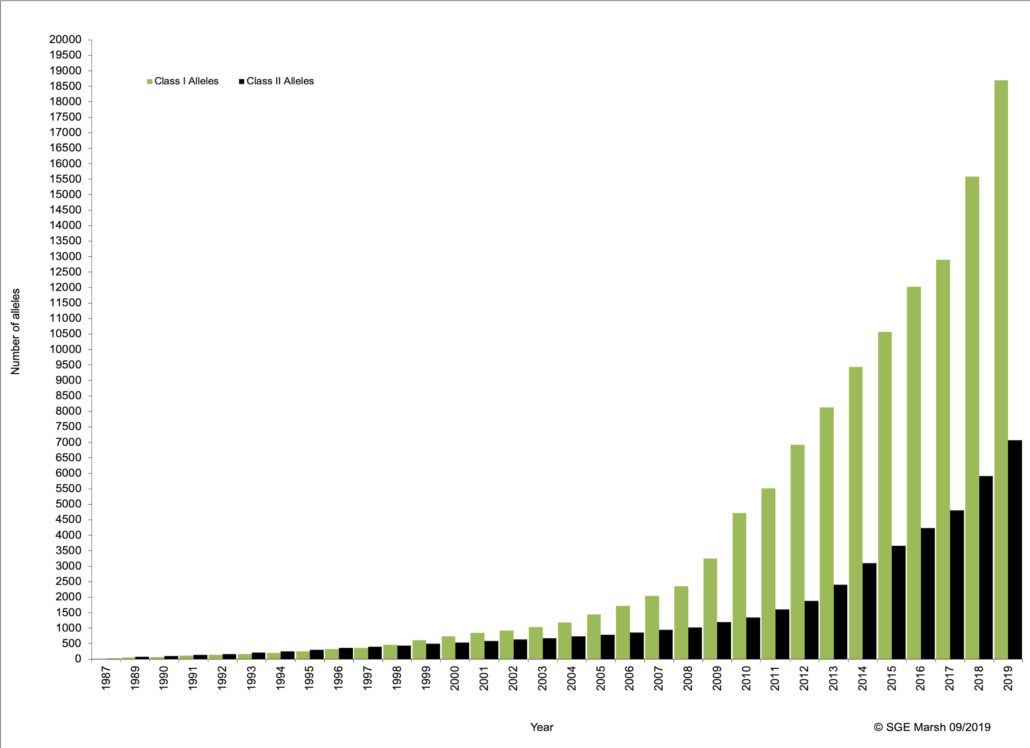
Fig. 1
The number of sequence depositions in the database is growing at an exponential rate (Fig. 1). The sequence count currently exceeds 25,000 and shows no signs of slowing down. Each version release includes a list of newly deposited sequences and allele reports.
HLA Nomenclature
The IMGT database reference pages include descriptions of the HLA nomenclature used in the database itself. We describe the interpretation of HLA nomenclature in our HLA reports and other KB articles. Due to the long history of HLA typing, the HLA nomenclature is fairly complex and can be rather difficult to interpret. However, IMGT includes documentation on this topic:
- Allele naming conventions
- Nomenclature reports
- Nomenclature updates
- HLA gene names
- HLA antigens
- HLA suffixes
- Class I alleles
- Class II alleles
Alleles by Another Name
One important note about HLA nomenclature is the unusual definition of “allele”. In contrast to our usual notion of genetic alleles (i.e. a single nucleotide polymorphism), within an HLA Typing field, specific and unique genomic sequences are referred to as alleles. For example, an HLA allele can refer to a single SNP, sets of SNP’s, single indels, sets of indels, or some combination of SNP’s and indels (i.e. a single substitution, insertion or deletion, or multiple substitutions, insertions, or deletions). Thus, each HLA “allele” is a unique genomic sequence that differs by at least one base pair from another HLA “allele”. One should keep in mind this distinction between conventional alleles and HLA alleles when working with the IMGT HLA database.