Discovery of a Recombination Event
In a recent HLA Next Gen sequencing run, one of our client’s samples displayed some puzzling results. A standard HLA typing report showed a near-perfect sequence alignment across the first two exons (and first intron) in allele HLA-DPA1 followed by a significant number of sequence read mismatches for everything downstream from exon 2. This is a highly unusual pattern that triggered a more in-depth analysis of this sample dataset.
In cases like this we first do some quality control checks to make sure there were no issues with DNA extraction, the library prep protocol and the (Illumina) sequencing instrument and sequencing run itself. We first checked the quality metrics for the sequence alignments and found them to be very good with high read depth, low noise levels, high estimated second allele percentages and good signal-to-noise ratio. Likewise, the Illumina sequencer run metrics were also very good with optimal cluster density, high Phred Q-scores, high percent reads passing filter and a host of other metrics. We concluded that the DNA extraction steps, execution of the library prep protocol and sequencing itself were likely done correctly.
We use GenDx NGSengine for HLA Typing of Next Gen Sequencing results. NGSengine performs sequence alignments of all sample reads against the IMGT/HLA database to find the most likely matching genotype and alleles. In this sample, NGSengine identified the best matching genotype as DPA1*01:03:01:04 / DPA1*01:05, however, it also included a fairly high number of exon and intron mismatches.
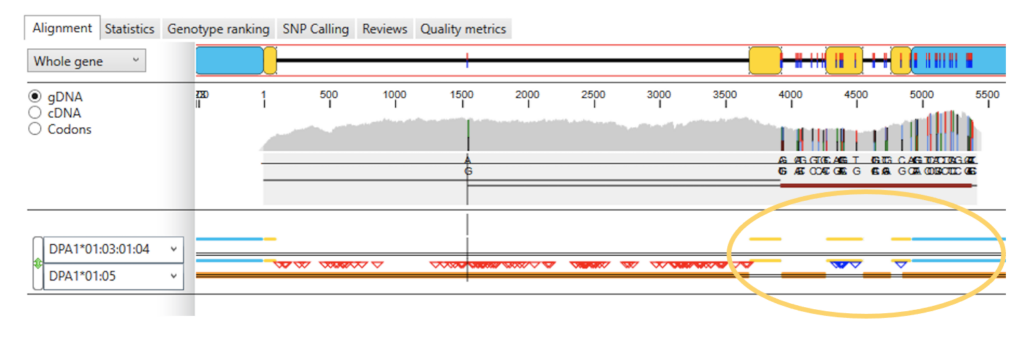
Fig. 1. Numerous exon and intron mismatches downstream from the end of exon 2 in allele DPA1*01:05 (blue triangles).
As shown in Fig. 1, there are no exon mismatches in exon 1, only a single mismatch in intron 1 and no exon mismatches in exon 2. However, there is a significant number of intron mismatches in introns 2 and 3 and there is a significant number of exon mismatches in exons 3 and 4 and also mismatches in the 3′-UTR region. In fact, there were 7 total exon mismatches and 112 total intron mismatches compared to the reference genotype from the IMGT/HLA database. This is a fairly high number of exon/intron mismatches, which merits further examination. (For our purposes here we’ll skip some other information present in Fig. 1).
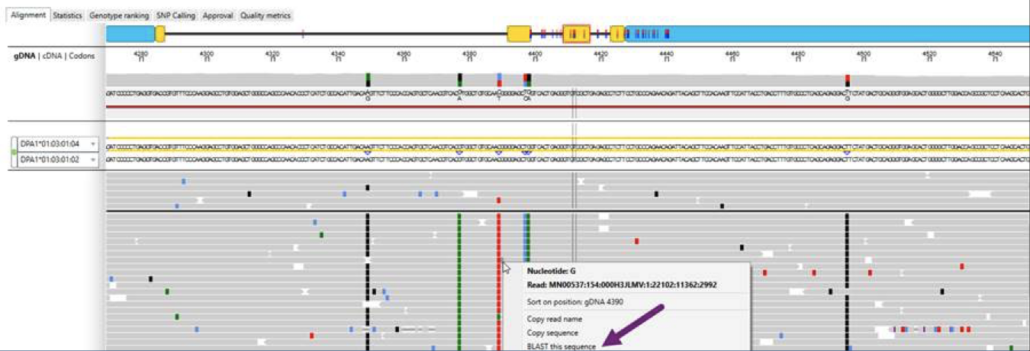
To facilitate deep dives into HLA typing datasets, NGSengine includes a feature that allows one to pick individual reads from the full set of reads and BLAST those reads individually against the IMGT/HLA database (see Fig. 2). Using this feature one can sometimes find reads that map to a different allele than the one initially identified by the default typing algorithm.
Fig. 2. BLAST’ing individual reads against the IMGT/HLA database.
If we choose some reads from the region before and including exon 2 and BLAST them against the IMGT/HLA database, we find that the reads map to DPA1*01:03:01:04, as might be expected from Fig. 1.
Fig. 3. BLAST results identify a different allele from the initial typing algorithm.
However, if we choose reads downstream from the end of exon 2, i.e. intron 2 and beyond, and BLAST them against the database, we find that the reads map to DPA1*02:01:01 (see Fig. 3). We would not expect this based on the results from the initial typing algorithm.
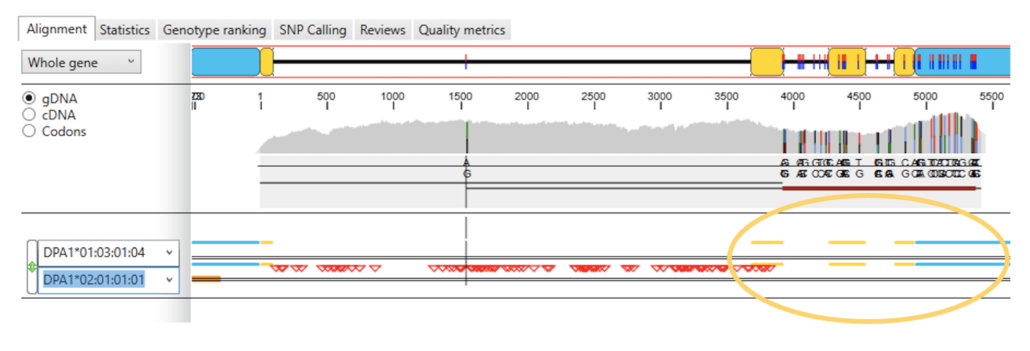
Fig. 4. No exon and intron mismatches downstream from the end of exon 2 in allele DPA1*02:01:01:01 (no blue triangles).
Manually substituting allele DPA1*02:01:01:01 for allele DPA1*01:05 (see Fig. 4), we see that all of the exon and intron mismatches disappear in the sequence alignment downstream from the end of exon 2, suggesting that sample reads at this genomic location belong to allele DPA1*02:01:01:01.
Considering the sequence alignment patterns above, we conclude that this sample contains a novel allele derived from a recombination event between DPA1*01:03:01:04 and DPA1*02:01:01:01. Apparently, exons 1 and 2 and intron 1 from DPA1*01:03:01:04 recombined with exons 3 and 4 and introns 2 and 3 from DPA1*02:01:01:01 to create a new allele. The significance of this specific recombination event is unknown but genetic recombination has been implicated in a variety of disease states and thus merits further investigation.