HLA-A*01:01:01:01 Allele and its Impact on the Development of Malignancies
The Human Leukocyte Antigen (HLA) system plays a crucial role in immune recognition and response against pathogens and malignant cells. The HLA-A*01:01:01:01 allele has been linked to an increased risk of developing certain malignancies, such as cervical cancer and non-small cell lung cancer (NSCLC). The current hypothesis suggests that this increased risk may be attributed to the HLA-A*01:01:01:01 molecule’s ability to present tumor-associated antigens (TAAs) to CD8+ T cells, thereby modulating the immune response against cancer cells. This paper aims to explore this hypothesis by delving into the molecular mechanisms of HLA-A*01:01:01:01-mediated antigen presentation, the role of CD8+ T cells in tumor surveillance, and potential implications for cancer immunotherapy.
Introduction
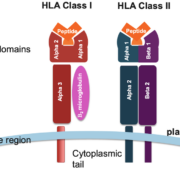
The HLA system, encoded by the Major Histocompatibility Complex (MHC) genes, is a set of cell surface proteins responsible for presenting antigenic peptides to T cells, thereby initiating adaptive immune responses. Class I MHC molecules, including HLA-A, HLA-B, and HLA-C, present endogenous antigens to CD8+ T cells, which play a vital role in immune surveillance against viral infections and malignancies. Certain HLA alleles have been associated with an increased risk of developing specific cancers, suggesting a role for these alleles in shaping the host immune response against cancer cells.
One such allele is the HLA-A*01:01:01:01, which has been linked to an increased risk of developing cervical cancer and NSCLC. It has been hypothesized that the HLA-A*01:01:01:01 molecule may present TAAs to CD8+ T cells more efficiently, thereby modulating the immune response against cancer cells. In this paper, we aim to explore the molecular mechanisms underlying this hypothesis, focusing on the role of HLA-A*01:01:01:01 in antigen presentation, CD8+ T cell function, and the potential implications for cancer immunotherapy.
HLA-A*01:01:01:01 Allele and Antigen Presentation
The HLA-A01:01:01:01 allele is characterized by specific amino acid residues within its peptide-binding groove, which may influence its ability to bind and present TAAs to CD8+ T cells. Several studies have demonstrated that the HLA-A*01:01:01:01 molecule can efficiently present a wide range of TAAs, including those derived from cervical cancer and NSCLC cell lines. In addition, the HLA-A*01:01:01:01 allele has been shown to preferentially bind peptides with hydrophobic anchor residues, which are often found in TAAs. This unique binding specificity may allow the HLA-A*01:01:01:01 molecule to efficiently present TAAs to CD8+ T cells, thereby modulating the host immune response against cancer cells.
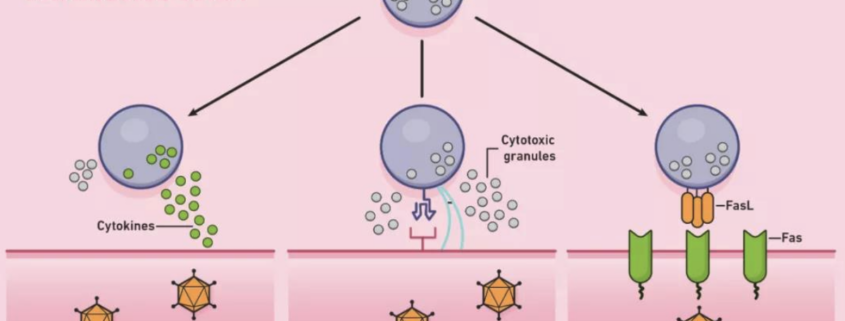
CD8+ T Cells and Tumor Surveillance
CD8+ T cells, also known as cytotoxic T lymphocytes (CTLs), play a crucial role in immune surveillance against cancer cells. Upon recognition of antigenic peptides presented by MHC class I molecules on the surface of cancer cells, CD8+ T cells become activated and exert their cytotoxic function through the secretion of perforin and granzymes, leading to target cell lysis. In the context of HLA-A*01:01:01:01-associated malignancies, it has been proposed that the efficient presentation of TAAs by the HLA-A*01:01:01:01 molecule may enhance the activation and cytotoxic function of CD8+ T cells against cancer cells. However, this increased activation may also result in an increased likelihood of immune escape mechanisms being employed by cancer cells, such as downregulation of MHC class I expression, secretion of immunosuppressive cytokines, and induction of regulatory T cells. As a result, the immune response against cancer cells may be modulated, leading to an increased risk of cancer development in individuals carrying the HLA-A*01:01:01:01 allele.
Potential Implications for Cancer Immunotherapy
The potential role of the HLA-A*01:01:01:01 allele in modulating the immune response against cancer cells has important implications for cancer immunotherapy. Firstly, understanding the molecular mechanisms of HLA-A*01:01:01:01-mediated antigen presentation and CD8+ T cell activation may provide insights into the development of personalized cancer vaccines targeting specific HLA-A*01:01:01:01-restricted TAAs. Secondly, the increased activation of CD8+ T cells in HLA-A*01:01:01:01-positive individuals may render these patients more responsive to immune checkpoint inhibitors, which aim to enhance T cell-mediated anti-tumor immunity. Finally, the identification of HLA-A*01:01:01:01-restricted TAAs may contribute to the development of T cell receptor (TCR)-engineered T cell therapies, in which T cells are genetically modified to express TCRs specific for tumor antigens, thereby enhancing their cytotoxic function against cancer cells.
Conclusion
The HLA-A*01:01:01:01 allele has been linked to an increased risk of developing certain malignancies, such as cervical cancer and NSCLC. The current hypothesis suggests that this increased risk may be attributed to the HLA-A*01:01:01:01 molecule’s ability to efficiently present TAAs to CD8+ T cells, thereby modulating the immune response against cancer cells. By exploring the molecular mechanisms of HLA-A*01:01:01:01-mediated antigen presentation and the role of CD8+ T cells in tumor surveillance, this paper has shed light on the potential implications for cancer immunotherapy. Further research is warranted to fully elucidate the role of HLA-A*01:01:01:01 in cancer development and to translate these findings into effective therapeutic strategies for patients with HLA-A*01:01:01:01-associated malignancies.
References
- Neefjes J, Jongsma MLM, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11(12):823-836.
- Gourraud PA, Khankhanian P, Cereb N, Yang SY, Feolo M, Maiers M, et al. HLA diversity in the 1000 genomes dataset. PLoS One. 2014;9(7):e97282.
- Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol. 1995;13:587-622.
- Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Sakaguchi K, Appella E, et al. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci U S A. 1994;91(14):6458-6462.
- Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213-219.
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69-74.
- Neller MA, López JA, Schmidt CW. Antigens for cancer immunotherapy. Semin Immunol. 2008;20(5):286-295.
- Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;35 Suppl:S185-S198.
- Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217-221.
- Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275-287.
- Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348(6230):62-68.


 The Sequencing Center
The Sequencing Center

Leave a Reply
Want to join the discussion?Feel free to contribute!