Cancer Gene Databases You Need
With any research, it all starts with the data. Cancer is no exception here. The incredible amount of funding that has been pushed into cancer research has produced many different databases that have identified genes that correlate with common cancers. A great first step for researchers exploring the genetic structures of these diseases is to compare their sequencing dataset against multiple cancer-based datasets. Historically, this has been a challenge simply because the datasets are stored all over the place, don’t communicate, and are hard to find. We’ve gone ahead and collected the 7 most commonly used cancer databases. Enjoy!
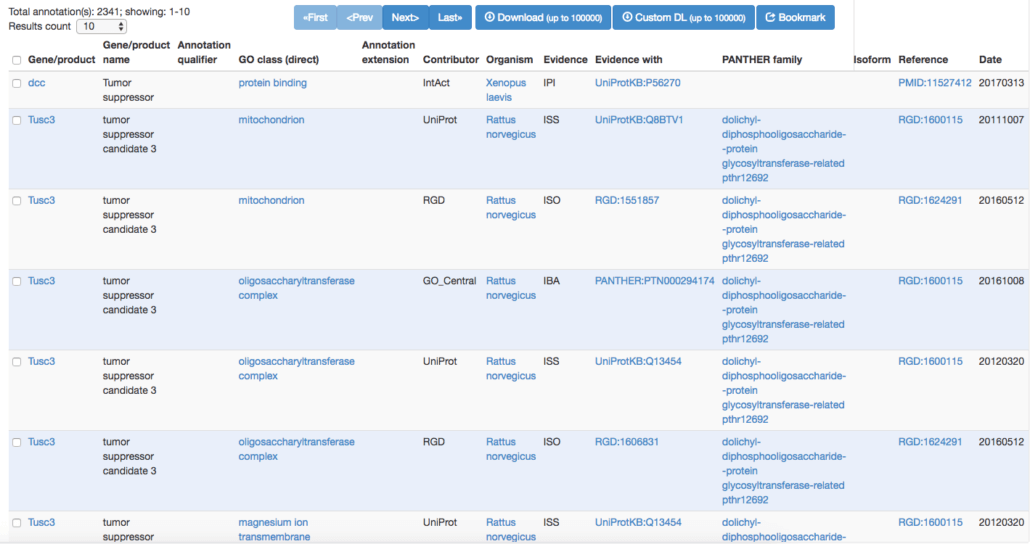
1. Gene Ontology Consortium
The Gene Ontology Consortium isn’t necessarily designed specifically for cancer genes, however, it does have an extensive amount of gene annotations. Using the search term “Tumor Suppressor“, you can scope in on gene annotations that have been associated with cancer. This portal is great for users who don’t necessarily need to do comparative genomics but rather want to scope in on suspected individual genes that may be contributing to the disease.
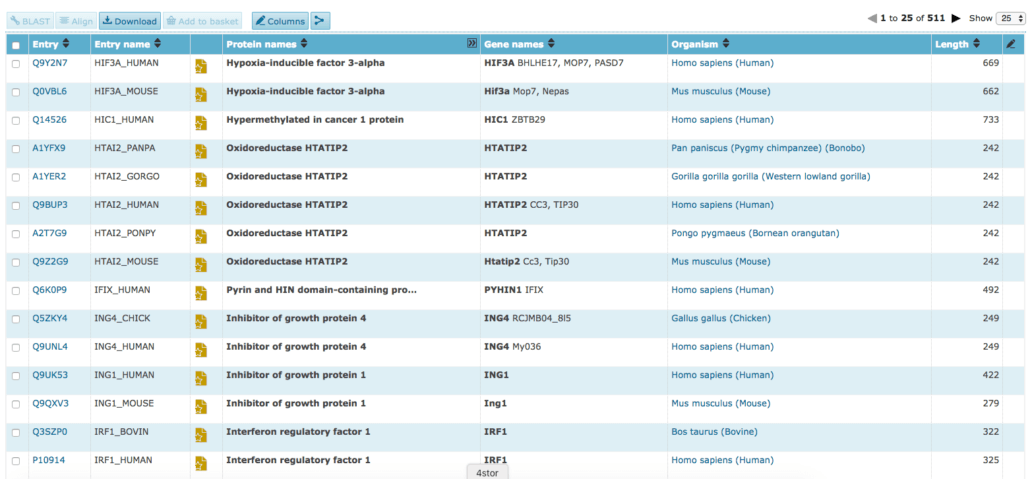
2. UniProtKB
The UniProtKB database is another great database that isn’t necessarily designed solely for cancer but has great searching mechanisms and tags built in place enabling users to easily search for these genes. By performing the search term “Tumor Suppressor”, you can get a healthy dataset of 500+ annotated genes in different species. What’s great about this database is that you can easily scope in through different filters as well as flexible columns, providing quick access to more insights on each row.
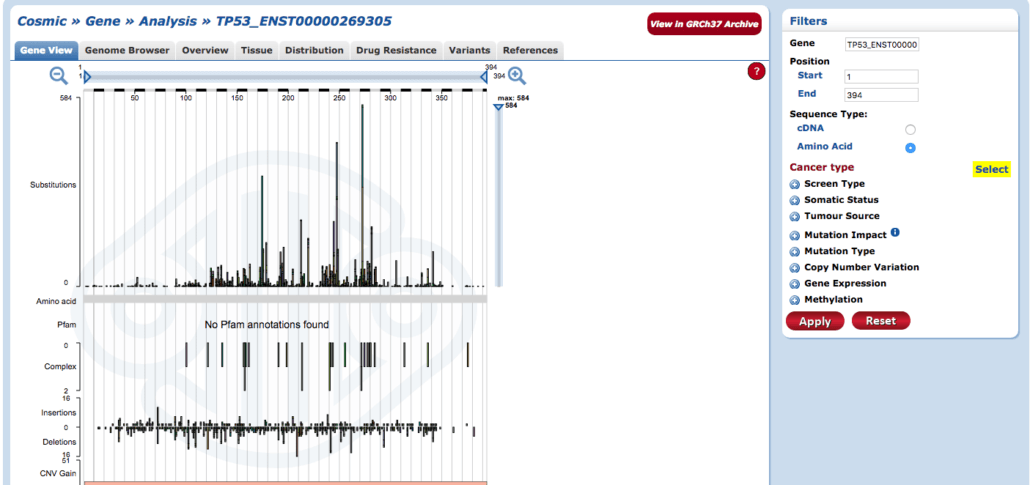
3. Cosmic: Catalogue of Somatic Mutations
Cosmic is one of our favorites when it comes to cancer gene databases. It provides great gene annotations, robust searching capabilities, great data visualizations, and arguably the best reference links & datasets out of all of them. Within a couple clicks and searches, we often find ourselves gathering more insights that we would have on other databases. Unfortunately, it doesn’t allow data uploading for comparison but don’t let that deter you. If you have an assembled genome and the variant file, you can quickly search for the common cancer gene suspects. If you don’t have either of those files, you should get in touch with us!
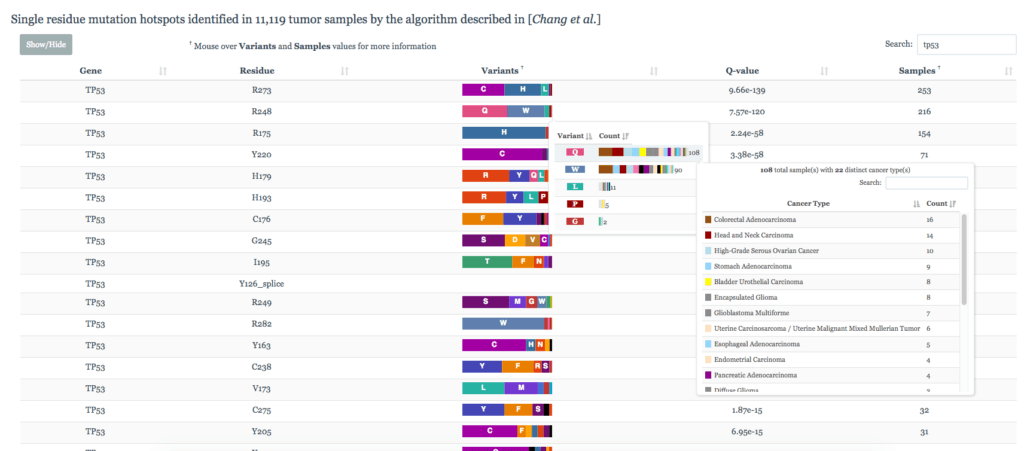
4. Cancer Hotspots
Hotspots is a database managed by Memorial Sloan Kettering. The database is full of mutations and cancer genes that they have deemed to meet the statistical significance threshold for being found in these types of diseases. It provides the fastest searching out of all of the databases and gives a great breakdown of variants found in what types of diseases. This is an effective tool for clinical researchers in the discovery and analysis process.
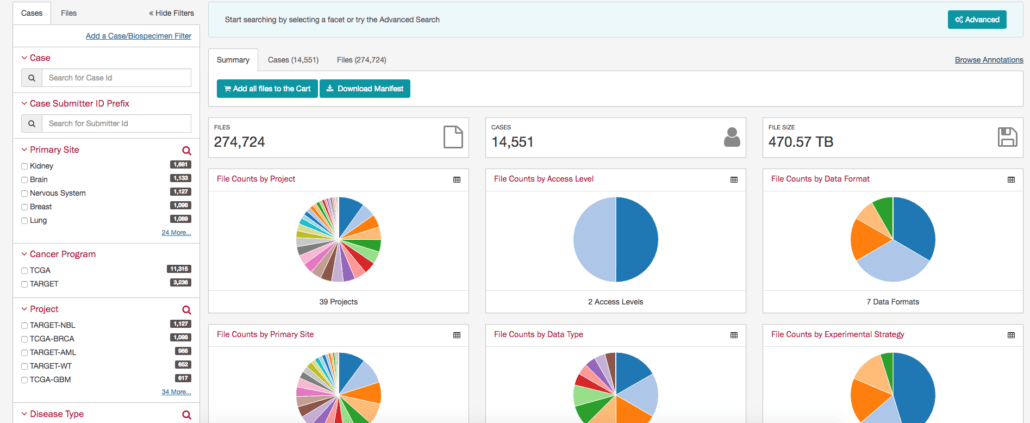
5. Harmonized Cancer Datasets – GDC Data Portal
Provided by the NCI, the GDC Portal provides nearly 5PB of cancer-related data. It has a unique filtering capability where you can filter by the primary site which makes it another highly effective tool for clinical researchers. With a simple click of checkboxes, I can filter my dataset down by gender, age, cancer location, race, and more. The only tricky thing about this site is that it’s primarily for downloading data into your own environment. If you want to do an analysis on the web, then you need to go to the GDC cBioPortal which allows you to retrieve back useful data visualizations.
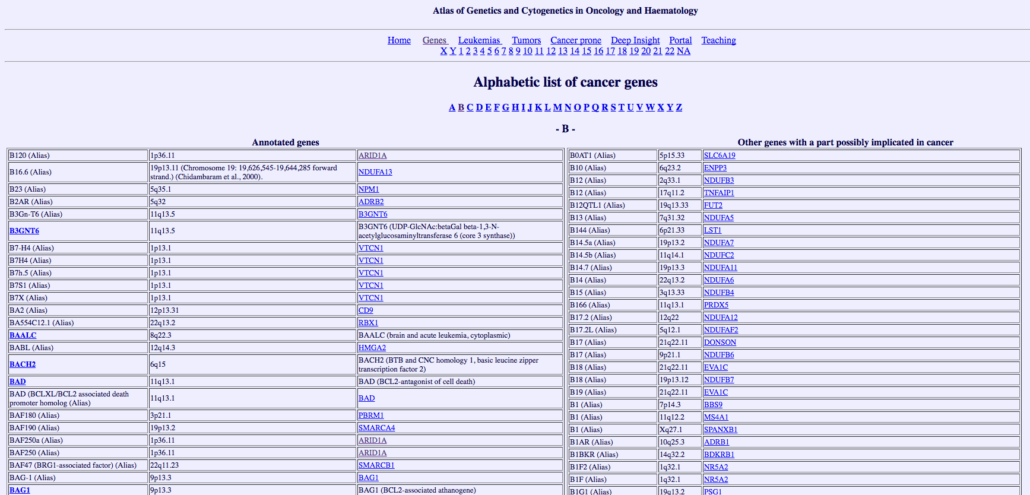
6. Atlas of Genetics and Cytogenetics in Oncology and Haematology
This list would not be complete without Atlas. Atlas doesn’t have a user-friendly interface and it can feel pretty old school but it has some of the richest cancer gene data out there. It provides robust research information on effectively all cases recorded. We use this tool as more of a research and exploration tool rather than a comparative dataset tool. With its massive gene dataset and example, it’s a website that deserves a place in your bookmarks.




Leave a Reply
Want to join the discussion?Feel free to contribute!